We have always been interested in the development of next generation genomic and proteomic technologies for studying living systems. Beyond the novel bioinformatics tools we work on and the proteomics methods we are innovating, our group is also active in the application of state of the art methods to better understand the molecules that are found in living systems.
Proteomics
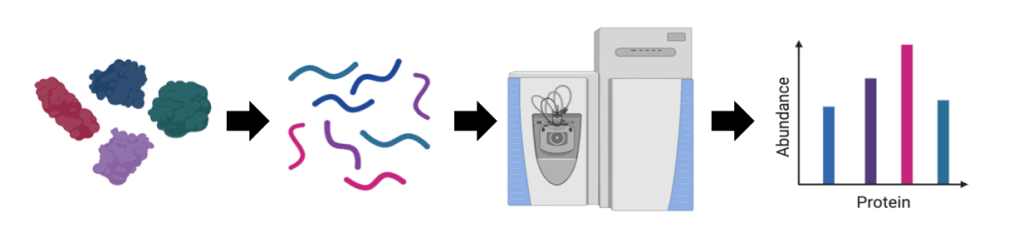
Mass spectrometry (MS) is an analytical tool for measuring the mass to charge ratio of molecules. Our laboratory has access to a suite of state of the art mass spectrometers to collect information about protein mixtures. While the application of existing technology is critical to our work, we are also interested in the development of next generation tools. We use high resolution mass spectrometry (HR-MS) coupled with liquid chromatography (LC) for such proteomics studies. Depending on the goals of a particular experiment, we do targeted proteomics and discovery proteomics with Data Dependent Acquisition (DDA) and Data Independent Acquisition (DIA). One important application is the use of DIA methods to quantify residual host cell proteins (HCPs) in biotherapeutics with label-free quantification, as HCPs can affect therapeutic safety and efficacy. Therefore, HCP levels need to be tightly controlled. HCPs are traditionally quantified with ELISA, which provides information on HCPs as a population. As an orthogonal approach to ELISA, LC-MS/MS identifies and quantifies each individual HCP and serves as a useful tool to support downstream process development. We are applying these methods to improve mAb and AAV production processes.
Genomics

Our work in the genomics space has been part of an international consortium of Chinese hamster ovary (CHO) researchers that we led which resulted in significant accomplishments including the sequencing and assembly of CHO reference genomes (www.CHOgenome.org). From this improved genome, we studied changes in the epigenome and transcriptome that occur in response to various bioprocess-related stresses and identified genetic hotspots that can be leveraged for therapeutic protein production.