Applied proteomics
In other work, we developed numerous methods in applied proteomics including:
- Development of antemortem diagnostic tests for prion disease and Alzheimer’s disease
- Characterization of drug interactions and transport of large molecules across the blood-brain barrier
- Creation of microfluidic nanoscale devices for improved protein separation and analysis
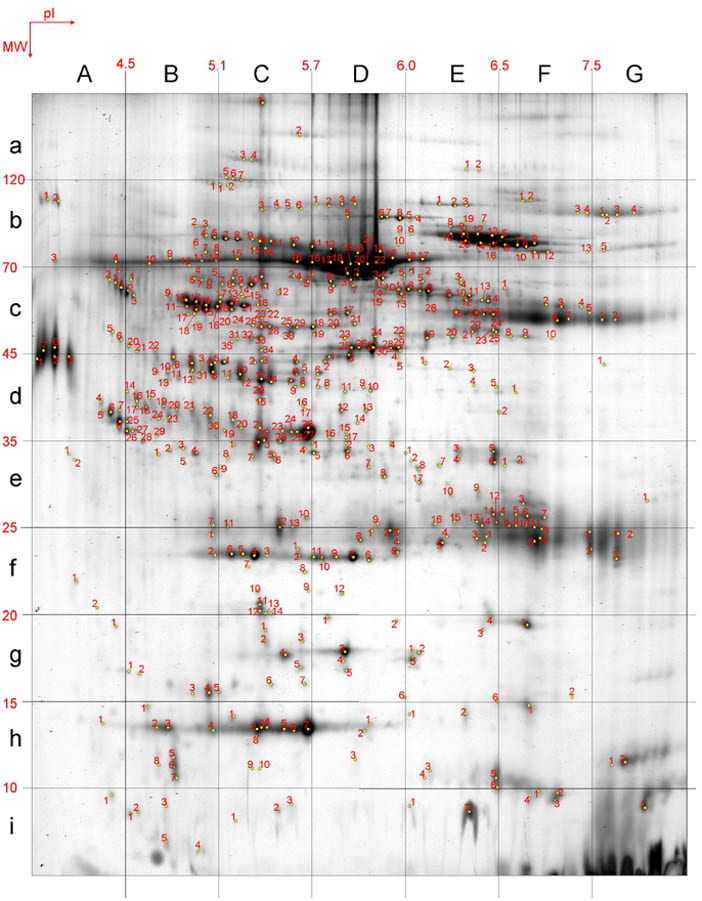
Figure 1: Hydrocephalus sample (Human Cerebrospinal Fluid (CSF) from Hydrocephalus Patient; 3-10NL; 12-15%T)
- We were the first academic laboratory to employ tandem time of flight mass spectrometry (MALDI TOF-TOF) and have published seminal papers demonstrating the use of 4plex and 8plex isobaric tagging methods for protein quantification.
- We applied proteomics to study biomarkers present in cerebrospinal fluid (CSF).
- We developed the first antemortem diagnostic test for Creutzfeldt-Jakob disease (CJD). CJD is one of a group of prion diseases (which includes “Mad Cow disease”) and in humans, this disease was previously only definitively diagnosed upon autopsy. We helped identify a protein marker in cerebrospinal fluid called 14-3-3 that is used to help evaluate patients suspected of having prion disease.
- We were also involved in the development of the first antemortem diagnostic tests for Azheimer’s disease. By studying a large group of known Alzheimer cases and other cases, we found a panel of protein markers that was effective in separating these two groups. In this work, we also characterized CSF in subjects undergoing new immunotherapies for Alzheimer’s disease to determine the efficacy of treatment.
Bioinformatics and systems biology
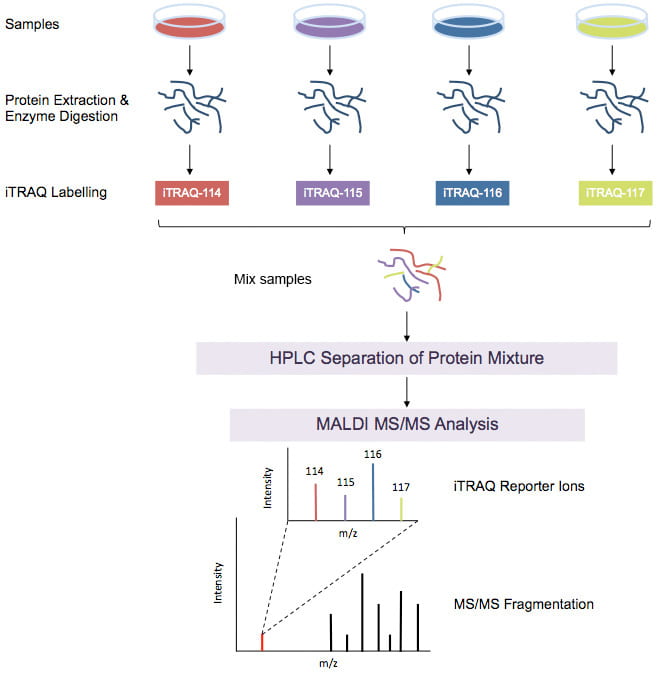
We also made notable scientific contributions in the fields of bioinformatics and computational systems biology. Bioinformatics efforts intersect computer science with life science and computational systems biology efforts intersect applied mathematics with life science. For example, we developed bioinformatic tools that facilitate the analysis of shotgun proteomics datasets such as those obtained using isobaric tagging reagents. We created a package called iTRAQPak which facilitates visualization of such datasets and addresses important issues related to protein versus peptide measurements during large scale shotgun proteomics studies. We also developed a novel modeling framework rooted in genome reconstruction to predict therapeutic protein glycan patterns of non-template driven glycosylation process with high accuracy.
Note: Data from the CSF or P. syringae 2DE maps and iTRAQPak tool is available upon request. Please email Kelvin Lee if interested.